Visualising Read Alignments on UCSC Genome¶
UCSC genome browser¶
UCSC genome browser is a web-based tool that includes a genome browser and sequence alignment tools. The genome browser supports many different file formats including BAM format. For more information please go to http://genome.ucsc.edu.
Setup Apache web server¶
In order create and view the custom track of our data, you can upload your file to UCSC web server. However, if you file is big, it will take a long time to upload. In this tutorial, we will set up Apache webserver and let UCSC genome browser download data from the amazon machine via HTTP.
First, install Apache using apt-get:
apt-get -y install apache2
/etc/init.d/apache2 start
Then use your internet browser to go to http://$amazon-machine-name, for example:
http://ec2-184-72-90-214.compute-1.amazonaws.com
You should see “It works!!”. That means it works.
Visualize BWA output¶
We will first look at the mapping result from BWA. The mapping result is saved in BAM format. We have to sort and index the file before visualize it on UCSC genome browser.
To sort and index BAM file, execute:
cd /data/drosophila
samtools sort RAL357_full_bwa.bam RAL357_full_bwa.sorted
samtools index RAL357_full_bwa.sorted.bam
Next, create links of our files to /var/www/:
ln -fs /data/drosophila/RAL357_full_bwa.sorted.bam /var/www/
ln -fs /data/drosophila/RAL357_full_bwa.sorted.bam.bai /var/www/
Create a custom track on UCSC genome browser¶
Using your internet browser to go to http://genome.ucsc.edu.
At the home page, click “Genomes” on the left corner.
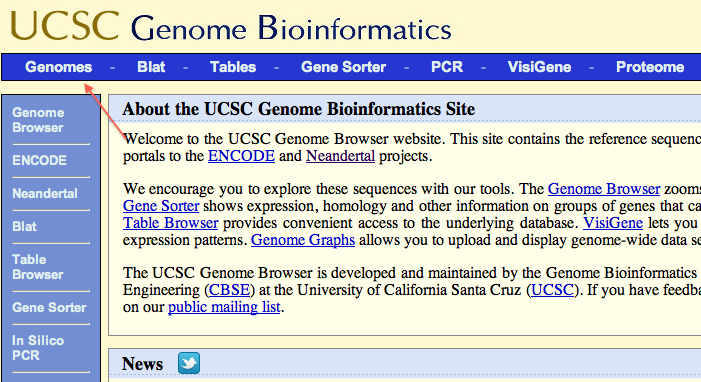
On the genomes page, select an organism of your chioce. In this case, select “D. melanogaster” from group “Insect”. Then click “manage custom track” or “add custom track” if you already have other custom tracks.
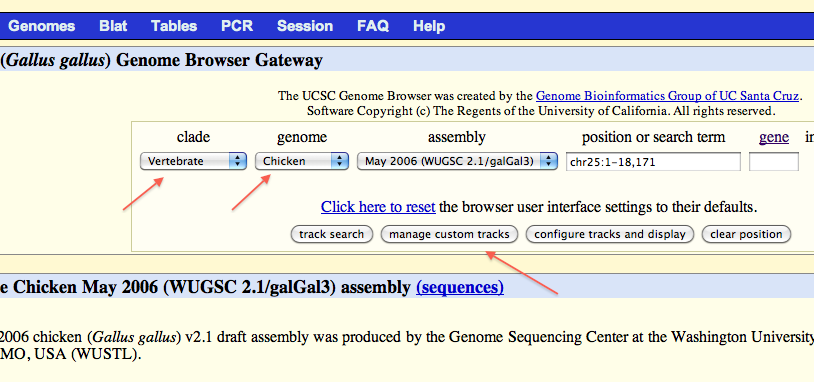
Copy and paste the line below in “Paste URLs or data:” box:
track name='bwa mapping' type=bam db=dm3 visibility=squish bigDataUrl=http://$amazon-machine-name/RAL357_full_bwa.sorted.bam
# name = track name (has to be unique) [default:User Track]
# type = file type (bam, bed, bigBed, wig, bedWig)
# db = genome
# visibility = display mode (full, squish, pack, dense)
# bigDataUrl = URL to a data file
# description = description [optional]
Don’t forget to change “$amazon-machine-name” to your amazon machine DNS.
Then click submit and click “Go to genome browser”. You should see something like this.
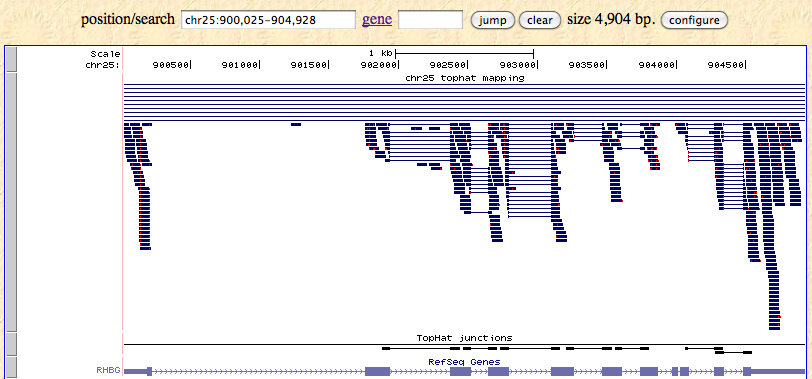
Adding a custom track using a URL is suitable and required for a large file. Large files need to be indexed by a tool downloadable from UCSC genome browser.
For a small file, you can upload a file directly from you local computer and it doesn’t need to be indexed.